Protein-coding gene in the species Homo sapiens
| CTSZ |
|---|
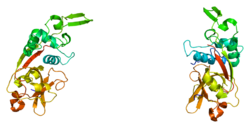 |
| Available structures |
|---|
| PDB | Ortholog search: PDBe RCSB |
|---|
| List of PDB id codes |
|---|
1DEU, 1EF7 |
|
|
| Identifiers |
|---|
| Aliases | CTSZ, CTSX, cathepsin Z |
|---|
| External IDs | OMIM: 603169; MGI: 1891190; HomoloGene: 1022; GeneCards: CTSZ; OMA:CTSZ - orthologs |
|---|
| Gene location (Human) |
|---|
 | | Chr. | Chromosome 20 (human)[1] |
|---|
| | Band | 20q13.32 | Start | 58,985,686 bp[1] |
|---|
| End | 59,008,238 bp[1] |
|---|
|
| Gene location (Mouse) |
|---|
 | | Chr. | Chromosome 2 (mouse)[2] |
|---|
| | Band | 2 H4|2 97.94 cM | Start | 174,269,286 bp[2] |
|---|
| End | 174,280,832 bp[2] |
|---|
|
| RNA expression pattern |
|---|
| Bgee | | Human | Mouse (ortholog) |
|---|
| Top expressed in | - monocyte
- granulocyte
- mucosa of transverse colon
- right coronary artery
- ascending aorta
- Descending thoracic aorta
- rectum
- upper lobe of left lung
- right lobe of liver
- right lung
|
| | Top expressed in | - stroma of bone marrow
- yolk sac
- dermis
- left lung lobe
- migratory enteric neural crest cell
- calvaria
- decidua
- supraoptic nucleus
- parotid gland
- efferent ductule
|
| | More reference expression data |
|
|---|
| BioGPS | 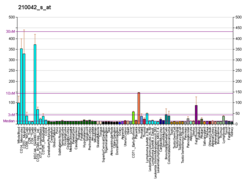 | | More reference expression data |
|
|---|
|
| Gene ontology |
|---|
| Molecular function | - protein binding
- peptidase activity
- hydrolase activity
- cysteine-type endopeptidase activity
- cysteine-type peptidase activity
- carboxypeptidase activity
| | Cellular component | - endoplasmic reticulum lumen
- COPII-coated ER to Golgi transport vesicle
- intracellular membrane-bounded organelle
- endoplasmic reticulum
- plasma membrane
- endoplasmic reticulum-Golgi intermediate compartment membrane
- extracellular exosome
- Golgi membrane
- extracellular region
- specific granule lumen
- ficolin-1-rich granule lumen
- extracellular space
- lysosome
- cell surface
- growth cone
- cytoplasmic vesicle
- cell cortex region
- collagen-containing extracellular matrix
| | Biological process | - proteolysis involved in cellular protein catabolic process
- COPII vesicle coating
- endoplasmic reticulum to Golgi vesicle-mediated transport
- epithelial tube branching involved in lung morphogenesis
- angiotensin maturation
- neutrophil degranulation
- proteolysis
- negative regulation of plasminogen activation
- negative regulation of neuron projection development
- negative regulation of protein binding
- positive regulation of neuron apoptotic process
- regulation of neuron death
- positive regulation of neural precursor cell proliferation
| | Sources:Amigo / QuickGO |
|
|
| Wikidata |
| View/Edit Human | View/Edit Mouse |
|
Cathepsin Z, also called cathepsin X or cathepsin P, is a protein that in humans is encoded by the CTSZ gene.[5][6] It is a member of the cysteine cathepsin family of cysteine proteases, which has 11 members.[7] As one of the 11 cathepsins, cathepsin Z contains distinctive features from others. Cathepsin Z has been reported involved in cancer malignancy and inflammation.
Structure
Gene
The CTSZ gene is located at 20q13.32 on chromosome 20, consisting of 6 exons. At least two transcript variants of this gene have been found, but the full-length nature of only one of them has been determined.[6]
Protein
Cathepsin Z is characterized by an unusual and unique 3-amino acid insertion in the highly conserved region between the glutamine of the putative oxynion hole and the active site cysteine. The pro-region of cathepsin Z shares no significant similarity with other cathepsin family sequences.[8] It contains only 41 amino acid residues without the conserved motif of ERFNIN or GNFD found in other cysteine proteinases. Besides, the proregion sequence contains no lysine residue.
Function
The protein encoded by this gene is a lysosomal cysteine proteinase and member of the peptidase C1 family. It exhibits both carboxy-monopeptidase and carboxy-dipeptidase activities. Up to date, eleven human cysteine proteinases have been identified, including cathepsin B, cathepsin C, cathepsin F, cathepsin H, cathepsin K, cathepsin L, cathepsin L2 or V, cathepsin O, cathepsin S, cathepsin Z, and cathepsin W. These cysteine proteinases belong to the papain family and represent a major component of the lysosomal proteolytic system. In addition to playing a critical role in protein degradation and turnover, these proteinases appear to play an extracellular role in a number of normal and pathological conditions. The human cathepsin Z contains distinctive features from other human cysteine proteases.[9] It is an exopeptidase with strict carboxypeptidase activity, while most other cathepsins are endopeptidases.[7] Cathepsin Z has an exposed integrin-binding Arg-Gly-Asp motif within the propeptide of the enzyme, through which cathepsin Z has been shown to interact with several integrins during normal homeostasis, immune processes and cancer.[10][11][12][13] It is also shown to bind cell surface heparin sulphate proteoglycans, indicating possible functions in cellular adhesion and phagocytosis.[14]
Clinical significance
This gene is expressed ubiquitously in cancer cell lines and primary tumors and, like other members of this family, may be involved in tumorigenesis. For instance, cathepsin Z promotes invasion and migration via a noncatalytic mechanism, suggesting multiple modes of cell invasion may be involved in cancer malignancy.[13] Cathepsin Z is also reported to have a protective, but not proteolytic, function in inflammatory gastric disease.[15] It is reported in another study that cathepsin Z may be responsible for dopamine neuron death and thus involved in the pathogenic cascade event.[16] Single-nucleotide polymorphism in CTSZ is found associated with tuberculosis susceptibility, indicating that the pathways involving this protein could yield novel therapies for tuberculosis.[17]
Interactions
Cathepsin Z has been shown to interact with the following proteins: CEP55, FBXO6, KIFC1, KRT40, KRTAP5-9, KRTAP5-9, LYPLAL1, MID2, MSN, MTUS2, NOTCH2NL, PLK2, PLSCR1, SGOL2, and SPRED2.[18]
Cathepsin Z has also been found to interact with:
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000101160 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000016256 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Santamaría I, Velasco G, Pendás AM, Fueyo A, López-Otín C (July 1998). "Cathepsin Z, a novel human cysteine proteinase with a short propeptide domain and a unique chromosomal location". The Journal of Biological Chemistry. 273 (27): 16816–23. doi:10.1074/jbc.273.27.16816. PMID 9642240.
- ^ a b "Entrez Gene: CTSZ cathepsin Z".
- ^ a b Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, Turk D (January 2012). "Cysteine cathepsins: from structure, function and regulation to new frontiers". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 1824 (1): 68–88. doi:10.1016/j.bbapap.2011.10.002. PMC 7105208. PMID 22024571.
- ^ Nägler DK, Zhang R, Tam W, Sulea T, Purisima EO, Ménard R (September 1999). "Human cathepsin X: A cysteine protease with unique carboxypeptidase activity". Biochemistry. 38 (39): 12648–54. doi:10.1021/bi991371z. PMID 10504234.
- ^ Nägler DK, Ménard R (August 1998). "Human cathepsin X: a novel cysteine protease of the papain family with a very short proregion and unique insertions". FEBS Letters. 434 (1–2): 135–9. doi:10.1016/s0014-5793(98)00964-8. PMID 9738465. S2CID 22899822.
- ^ Lechner AM, Assfalg-Machleidt I, Zahler S, Stoeckelhuber M, Machleidt W, Jochum M, Nägler DK (December 2006). "RGD-dependent binding of procathepsin X to integrin alphavbeta3 mediates cell-adhesive properties". The Journal of Biological Chemistry. 281 (51): 39588–97. doi:10.1074/jbc.M513439200. PMID 17065156.
- ^ Kos J, Jevnikar Z, Obermajer N (April–June 2009). "The role of cathepsin X in cell signaling". Cell Adhesion & Migration. 3 (2): 164–6. doi:10.4161/cam.3.2.7403. PMC 2679876. PMID 19262176.
- ^ Obermajer N, Svajger U, Bogyo M, Jeras M, Kos J (November 2008). "Maturation of dendritic cells depends on proteolytic cleavage by cathepsin X". Journal of Leukocyte Biology. 84 (5): 1306–15. doi:10.1189/jlb.0508285. PMC 3252843. PMID 18701767.
- ^ a b c Akkari L, Gocheva V, Kester JC, Hunter KE, Quick ML, Sevenich L, Wang HW, Peters C, Tang LH, Klimstra DS, Reinheckel T, Joyce JA (October 2014). "Distinct functions of macrophage-derived and cancer cell-derived cathepsin Z combine to promote tumor malignancy via interactions with the extracellular matrix". Genes & Development. 28 (19): 2134–50. doi:10.1101/gad.249599.114. PMC 4180975. PMID 25274726.
- ^ a b Teller A, Jechorek D, Hartig R, Adolf D, Reißig K, Roessner A, Franke S (January 2015). "Dysregulation of apoptotic signaling pathways by interaction of RPLP0 and cathepsin X/Z in gastric cancer". Pathology, Research and Practice. 211 (1): 62–70. doi:10.1016/j.prp.2014.09.005. PMID 25433997.
- ^ Krueger S, Bernhardt A, Kalinski T, Baldensperger M, Zeh M, Teller A, Adolf D, Reinheckel T, Roessner A, Kuester D (2013). "Induction of premalignant host responses by cathepsin x/z-deficiency in Helicobacter pylori-infected mice". PLOS ONE. 8 (7): e70242. Bibcode:2013PLoSO...870242K. doi:10.1371/journal.pone.0070242. PMC 3728094. PMID 23936173.
- ^ Pišlar AH, Zidar N, Kikelj D, Kos J (July 2014). "Cathepsin X promotes 6-hydroxydopamine-induced apoptosis of PC12 and SH-SY5Y cells". Neuropharmacology. 82: 121–31. doi:10.1016/j.neuropharm.2013.07.040. PMID 23958447. S2CID 22499368.
- ^ Adams LA, Möller M, Nebel A, Schreiber S, van der Merwe L, van Helden PD, Hoal EG (June 2011). "Polymorphisms in MC3R promoter and CTSZ 3'UTR are associated with tuberculosis susceptibility". European Journal of Human Genetics. 19 (6): 676–81. doi:10.1038/ejhg.2011.1. PMC 3110050. PMID 21368909.
- ^ "CTSZ interaction network". BioGRID. Retrieved 6 August 2016.
- ^ Hafner A, Glavan G, Obermajer N, Živin M, Schliebs R, Kos J (August 2013). "Neuroprotective role of γ-enolase in microglia in a mouse model of Alzheimer's disease is regulated by cathepsin X". Aging Cell. 12 (4): 604–14. doi:10.1111/acel.12093. PMID 23621429. S2CID 23835547.
Further reading
- Nägler DK, Ménard R (August 1998). "Human cathepsin X: a novel cysteine protease of the papain family with a very short proregion and unique insertions". FEBS Letters. 434 (1–2): 135–9. doi:10.1016/S0014-5793(98)00964-8. PMID 9738465. S2CID 22899822.
- Pungercar J, Ivanovski G (2000). "Identification and molecular cloning of cathepsin P, a novel human putative cysteine protease of the papain family". Pflügers Archiv. 439 (3 Suppl): R116–8. doi:10.1007/s004240000112. PMID 10653162. S2CID 22775842.
- Pungercar J, Viyjak A, Ivanovski G, Krizaj I (2000). "Tissue expression and immunolocalization of a novel human cathepsin P". Pflügers Archiv. 439 (3 Suppl): R119–21. doi:10.1007/s004240000113. PMID 10653163. S2CID 22728027.
- Sivaraman J, Nägler DK, Zhang R, Ménard R, Cygler M (January 2000). "Crystal structure of human procathepsin X: a cysteine protease with the proregion covalently linked to the active site cysteine". Journal of Molecular Biology. 295 (4): 939–51. doi:10.1006/jmbi.1999.3410. PMID 10656802.
- Guncar G, Klemencic I, Turk B, Turk V, Karaoglanovic-Carmona A, Juliano L, Turk D (March 2000). "Crystal structure of cathepsin X: a flip-flop of the ring of His23 allows carboxy-monopeptidase and carboxy-dipeptidase activity of the protease". Structure. 8 (3): 305–13. doi:10.1016/S0969-2126(00)00108-8. PMID 10745011.
- Deussing J, von Olshausen I, Peters C (April 2000). "Murine and human cathepsin Z: cDNA-cloning, characterization of the genes and chromosomal localization". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1491 (1–3): 93–106. doi:10.1016/s0167-4781(00)00021-x. PMID 10760573.
- Bonthron DT, Hayward BE, Moran V, Strain L (August 2000). "Characterization of TH1 and CTSZ, two non-imprinted genes downstream of GNAS1 in chromosome 20q13". Human Genetics. 107 (2): 165–75. doi:10.1007/s004390000344. PMID 11030415. S2CID 25570066.
- Hartley JL, Temple GF, Brasch MA (November 2000). "DNA cloning using in vitro site-specific recombination". Genome Research. 10 (11): 1788–95. doi:10.1101/gr.143000. PMC 310948. PMID 11076863.
- Simpson JC, Wellenreuther R, Poustka A, Pepperkok R, Wiemann S (September 2000). "Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing". EMBO Reports. 1 (3): 287–92. doi:10.1093/embo-reports/kvd058. PMC 1083732. PMID 11256614.
- Wiemann S, Arlt D, Huber W, Wellenreuther R, Schleeger S, Mehrle A, Bechtel S, Sauermann M, Korf U, Pepperkok R, Sültmann H, Poustka A (October 2004). "From ORFeome to biology: a functional genomics pipeline". Genome Research. 14 (10B): 2136–44. doi:10.1101/gr.2576704. PMC 528930. PMID 15489336.
- Puzer L, Barros NM, Oliveira V, Juliano MA, Lu G, Hassanein M, Juliano L, Mason RW, Carmona AK (March 2005). "Defining the substrate specificity of mouse cathepsin P". Archives of Biochemistry and Biophysics. 435 (1): 190–6. doi:10.1016/j.abb.2004.12.007. PMID 15680921.
- Mehrle A, Rosenfelder H, Schupp I, del Val C, Arlt D, Hahne F, Bechtel S, Simpson J, Hofmann O, Hide W, Glatting KH, Huber W, Pepperkok R, Poustka A, Wiemann S (January 2006). "The LIFEdb database in 2006". Nucleic Acids Research. 34 (Database issue): D415–8. doi:10.1093/nar/gkj139. PMC 1347501. PMID 16381901.
- Lechner AM, Assfalg-Machleidt I, Zahler S, Stoeckelhuber M, Machleidt W, Jochum M, Nägler DK (December 2006). "RGD-dependent binding of procathepsin X to integrin alphavbeta3 mediates cell-adhesive properties". The Journal of Biological Chemistry. 281 (51): 39588–97. doi:10.1074/jbc.M513439200. PMID 17065156.
External links
- The MEROPS online database for peptidases and their inhibitors: C01.013[permanent dead link]
- Overview of all the structural information available in the PDB for UniProt: Q9UBR2 (Cathepsin Z) at the PDBe-KB.
|
|---|
| Activity | |
|---|
| Regulation | |
|---|
| Classification | |
|---|
| Kinetics | |
|---|
| Types | |
|---|
Portal: Biology
Biology
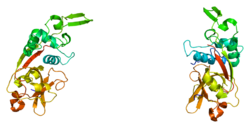
 1deu: CRYSTAL STRUCTURE OF HUMAN PROCATHEPSIN X: A CYSTEINE PROTEASE WITH THE PROREGION COVALENTLY LINKED TO THE ACTIVE SITE CYSTEINE
1deu: CRYSTAL STRUCTURE OF HUMAN PROCATHEPSIN X: A CYSTEINE PROTEASE WITH THE PROREGION COVALENTLY LINKED TO THE ACTIVE SITE CYSTEINE 1ef7: CRYSTAL STRUCTURE OF HUMAN CATHEPSIN X
1ef7: CRYSTAL STRUCTURE OF HUMAN CATHEPSIN X




















